Concept Search
Free Search
Classifier
Abstracts
Parameters
Vocabularies
All relations
Network
Abstracts
Concept set
Current Concepts
Saved Sets
Intra-set relations
Abstracts
Report parameters
Display mode
Current Set
Saved Sets
For questions, feature requests and other suggestions, please email us at kmine@tenwise.nl.
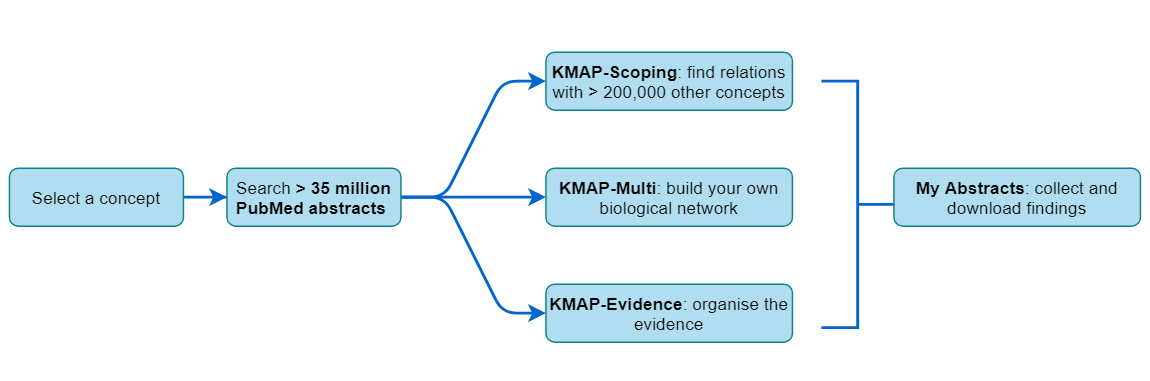
Concept search
The first tab on this page uses the Concept search method. You enter your concept of interest.A concept is a biological entity that can be:
- a bacterial gene
- a human gene
- a gene complex
- a metabolite
- a molecular pathway
- a phenotype
- a disease
- a drug
- an organism
Free search
The second tab uses the Free search method. This search method directly scans the 5000 most recent abstracts on your given input term, and then provides its results for further analyses.The results box displays navigation buttons to the Overview and Evidence tabs, and also refers to the more advanced tabs and methods available on the website.
This pages shows abstracts for your selected concept, ranked between 0 and 1 for various types of evidence. The score is determined by running a previously trained Random Forest model on the abstracts. Evidence is defined as the biological context of the abstract in which the concept appears. For example, an abstract with a score of 1 for clinical, is most likely about a clinical trial, or an observational study with patients. Likewise, an abstract with a high score for mouse is most likely about a mouse experimental model. The type of model can be selected in the left hand selection panel and described below. Not all models are available in the model if you are not logged in.
- Clinical: Clinical trials, patient observations
- Variant: Polymorphisms, DNA rearrangements, mutations
- 3R: Studies regarding 3R (Replacement, Reduction, and Refinement)
- Microbiome: Microbiota, metagenomics and 16S profiling studies
- Mouse: Experimental mouse models, studies on mice derived data
- Rat: Experimental rat models, studies on rat derived data
In all cases the 100 abstracts with the highest score for the selected model are shown. By changing the selection of the model, the output in the tab Evidence is automatically updated. The generated table (Article classes) has the following columns:
Citation: The articles in which your concept occurs selected for high scores with the selected model.
Score: The score (between 0 and 1) from the randomForest model
The search field button allow to select words used in the citations. This can be any word including author names and journal abbreviations.
Year: The year of the publication.
Clicking a row in the table shows the corresponding abstract on the righthand side. Clicking on the hyperlink (number) brings you to the abstract on the Pubmed site. Abstracts in the table can be selected and added to you set by pressing the button: Add selected abstracts to My Abstracts. If you go to My Abstracts then you will find the selected abstacts listed so you can revisit this at a later stage. The selection in My Abstracts does not change in case search conditions are altered.
On this advanced tab, the relations are shown between your concept and the vocabularies that are selected in the left panel. The total number of hits with your concept and the total of terms in the vocabulary are displayed in parenthesis.
Relations The table that is generated indicates per column:
- Name: Name of the concept. Clicking on the header sorts alphabetically.
- Type: The vocabulary from which the concept is derived.
- Nr. Abstracts: The number of abstracts in which both concepts co-occur.
- Score: A score denoting the uniqueness of the relation. THis is a score that takes into account the frequency of the individual terms and the frequency of the co-occurrence.
The Search field can be used to search in the entire table for a keyword. Selecting a row in the table shows the abstracts in the right results panel. Abstracts in the table can be selected by clicking one or more rows in this table and added to your Abstract Set by pressing the Add selected abstracts to Abstract Set. The collected abstracts can be visited anytime via the My Abstracts tab. The selection in My Set does not change in case search conditions are altered.
Clicking the button ‘Add Concept to KMAP Multi Set’ when a concept is selected in the table add this concept to your collected concept set which you can further explore using the KMAP-Multi tab.
Network This network shows the data from the Relations in a network with the selected concept in the center and up to a number of 50 other concepts per category. These concepts (dots with their name inside) have a different colour dependent on the type (same colors used as for the worldclouds ). In the righthand above corner a bar called ‘number of connections per category’ can be used to create and alter the network picture using between 0 and 50 concepts per category. The thickness of the arrows between the nodes indicate the relationship between the concepts. The thicker the node the higher the score on abstracts in which the two co-occur. By clicking on the arrow you are redirected to the TenWise server that depicts the abstracts with the co-occurrences. By clicking on the nodes you are redirected to the TenWise server that depicts the abstracts on this single concept. Finally the network shape can be changed manually by dragging the nodes across the screen. Screenshots can be made and used for reporting ed.
This tab shows analysis results for the set of concepts that have been saved in the concept set. This tab has a number of sub-tabs.
Concept set: The concepts that are currently in your set.
Relations: Visualization options to display how all concepts in your set are linked to each other. The network option displays the relations in a network in which each node is a concept. Clicking on a row in the table, or an edge in the network, displays the abstracts in which both concepts co-occur. This can also be done in sentence mode, in which case only the sentences in which both concepts co-occur are shown.
Report: A menu for you to create your own literature report. You can add various parameters, such as desired BioSets to be added to the relations found in the report (e.g. Chemokines, Lung Diseases, Kinases) and new relations that will be displayed between your concepts and selected vocabularies (e.g. Human Genes, Metabolites, Drugs).
On this page the references and their abstracts that are currently in the set, are shown. Using the radiobuttons on the left panel, you can format the output that you need. The default output is to show all the abstracts with citation information. If you need the identifiers, for example to import them into a reference manager program like EndNote, choose the PMIDS option. If you want to copy the abstracts directly into your document, select the Citations option.
You can use the download button to download the abstracts to an Excel file. This file contains all the information on authors, journal etc.
In other words, the score is calculated on basis of the number of abstracts in which both concepts co-occur and corrected for the number of abstracts in which both concepts occur alone, i.e. not with each other. This is also graphically explained in the figure below.
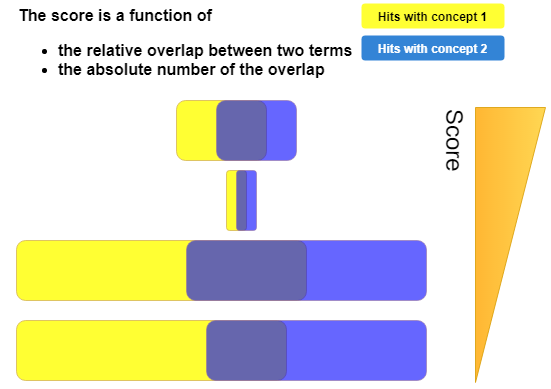
For a thorough description on how to score cooccurrences, we suggest to read the introductory chapter of the PhD thesis of Stefan Evert The Statistics of Word Cooccurrences.
- We want to provide a direct link from our text mining results to the underlying source. When we would run our textmining on full text papers, this becomes problematic if not all users have access to the same set of full text articles due to restrictions in subscriptions. This could solved by running only on the subset of articles that are full text, but compared to the entire MEDLINE database, this is a small set, and we may miss important relations.
-
Mining on full text on the one hand yields more relations, but at the same time also retrieves relations that are less informative. For example, relations that are retrieved from the introduction is often a repetition of known knowledge, whereas relations from the discussion section may represent speculations that are not always backed up by experimental evidence.
This does not mean that full text papers should not be used at all for text mining, but we believe that for this particular web application, abstracts provide a good trade-off between sensitivity (retrieving known facts) and specificity (discarding false positives).
More importantly, we believe that:
- Such weighing schemes will not significantly alter the biological relations that are observed in literature.
- Only biological experts can make the final call on whether a biological relation that is retrieved from literature is most useful for their particular research question, data analysis or hypothesis generation.
If you do not have login information, contact us for a trial account.